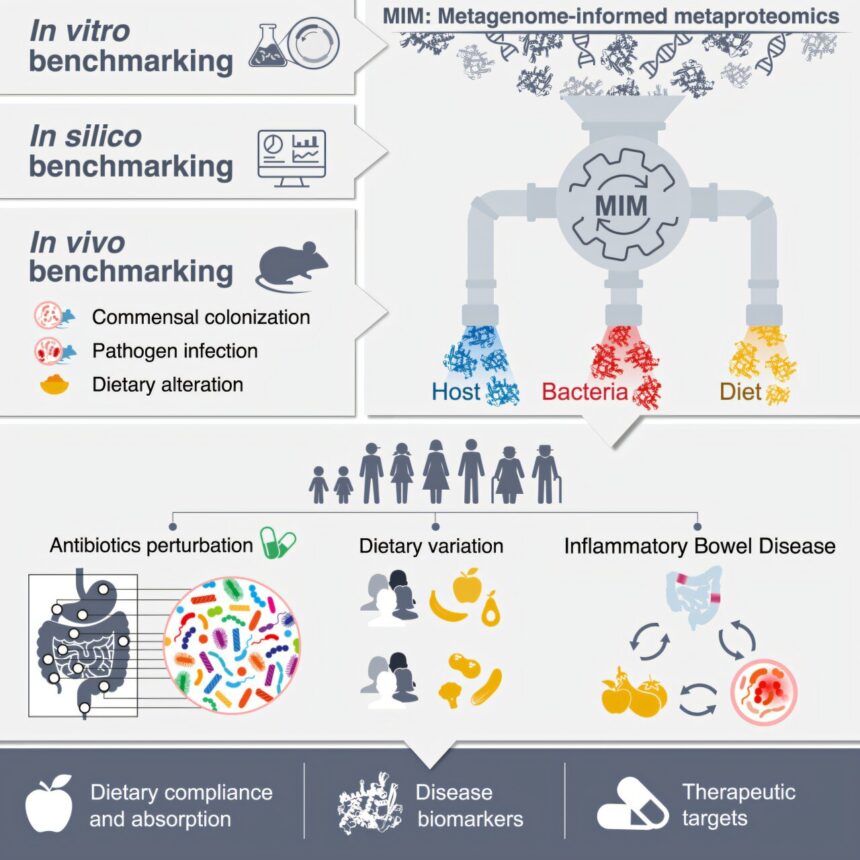
The intestines are often referred to as the “voice” of our bodies, capable of revealing hidden truths about our lifestyle and health. Researchers at the Weizmann Institute of Science have developed a groundbreaking method to decode the interactions among proteins in the intestine, shedding light on vital information for biomedical and clinical research.
Published in Cell, the study introduces a method that can identify all the proteins in the intestine, including those from food, the body, and the intestinal microbiome from a simple stool sample. This method, known as IPHOMED (Integrated Proteo-genomics of HOst, MicrobiomE, and Diet), offers unprecedented accuracy and resolution in understanding the complex protein interactions within the gut.
The researchers focused on the microbiome and its role in gut health. By combining DNA sequencing and mass spectrometry, they created a patient-specific protein map that reveals the activity of bacteria in the gut and the body’s response to these microorganisms. This approach uncovered previously unknown antimicrobial peptides secreted by the human gut, influencing the composition of the microbiome and individual susceptibility to disease.
Moreover, the Weizmann method has the unique ability to track an individual’s diet with remarkable precision. By analyzing proteins unique to different food items, the researchers could determine what a person had eaten based on stool samples. This noninvasive technique proved effective in identifying dietary habits, tracking changes in diet, and even detecting non-compliance with prescribed diets in patients with gastrointestinal diseases.
The application of this method to patients with inflammatory bowel disease yielded promising results. The analysis of stool samples provided detailed insights into the molecular interactions driving the disease, leading to the discovery of potential drug targets and new biomarkers for diagnosing and monitoring the condition. Additionally, the method demonstrated its utility in detecting diseases of the small intestine, offering a noninvasive approach to diagnosis.
Overall, the Weizmann Institute’s innovative method opens new possibilities for personalized nutritional and medical interventions in various disorders influenced by the microbiome. By deciphering the intricate language of intestinal proteins, researchers aim to develop tailored treatments for inflammatory, metabolic, malignant, and neurodegenerative diseases.
This groundbreaking research, published in Cell, marks a significant advancement in understanding intestinal health and disease. The Weizmann Institute of Science continues to lead the way in cutting-edge research that has the potential to revolutionize personalized medicine and improve patient outcomes.