The DTU National Food Institute, in collaboration with 11 European universities and institutions, has developed a groundbreaking method for analyzing data from wastewater monitoring. This innovative approach can help identify the sources of disease-causing bacteria, viruses, and antimicrobial resistance, whether they originate from humans, animals, industry, or the environment.
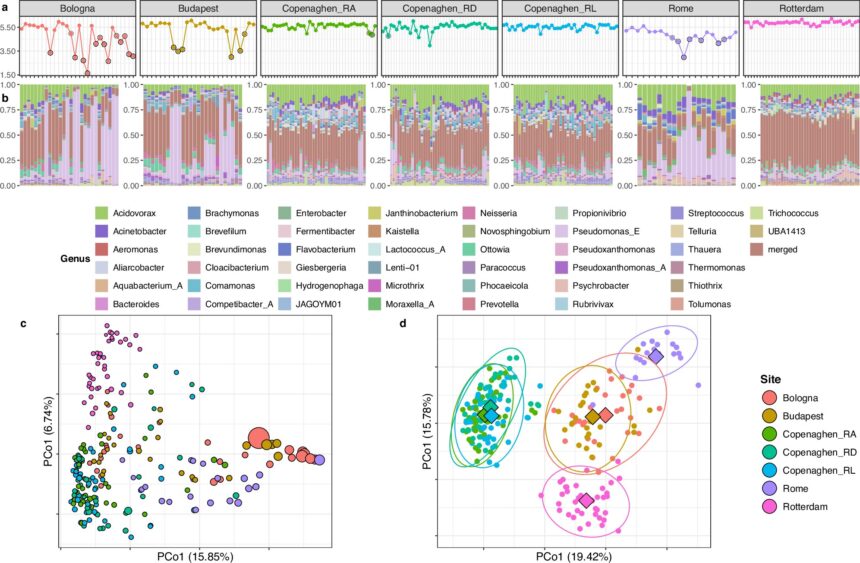
The research, recently published in Nature Communications, focuses on the abundance and diversity of recovered genomes in wastewater samples collected from seven treatment plants across five major European cities. The study highlights the potential of detecting multiple threats simultaneously, including antimicrobial resistance and cholera bacteria, which could play a crucial role in preventing disease outbreaks from escalating into epidemics.
Assistant Professor Patrick Munk, the corresponding author of the research paper, explains the challenges involved in extracting valuable data from wastewater. The samples contain a mix of known and unknown bacteria from various sources, making analysis complex. Additionally, the composition of wastewater can fluctuate due to seasonal changes in temperature.
To address these challenges, the researchers have developed a new computer program that enhances metagenomics-based wastewater monitoring. Unlike PCR testing, which screens for one threat at a time, this method can assess thousands of threats simultaneously. Professor Frank Aarestrup, a co-author of the study, emphasizes the value of historical data in enhancing the analysis of new samples.
The study’s findings are particularly relevant as European cities are mandated to monitor antimicrobial resistance in wastewater. In Denmark, the Statens Serum Institut is leading a large European collaboration to implement this monitoring system.
One surprising discovery from the study was the presence of cholera bacteria near the Avedøre Wastewater Treatment Plant in Copenhagen. Despite the minimal amounts found, the bacteria’s presence highlights the importance of continuous monitoring to prevent potential outbreaks.
The researchers also developed a software program to analyze the vast datasets generated from the wastewater samples. By clustering bacteria into distinct groups based on their behavior over time, the program can help identify the origins of specific species.
Metagenomics, a technique that analyzes mixed DNA from various microbial species, plays a crucial role in monitoring disease-causing bacteria and antibiotic resistance genes. The researchers identified over 1,300 previously unknown bacterial species in the wastewater samples, demonstrating the method’s effectiveness in detecting new threats.
Moving forward, the researchers aim to optimize the method further to improve its accuracy and success rate. By combining metagenomics-based wastewater surveillance with targeted PCR tests for specific threats, authorities can enhance disease surveillance and prevention efforts.
Overall, this research represents a significant step towards improving epidemic monitoring through wastewater analysis. The collaboration between European institutions underscores the importance of innovative approaches in public health surveillance and disease prevention.